This is the official implementation for cellNN, based on the work of “Cell Spatial Analysis in Crohn’s Disease: Unveiling Local CellArrangement Pattern with Graph-based Signatures”.
Source code repository
https://github.com/MASILab/cellNN
Get our docker image
docker pull onealbao/hovernet_gca:1.1
Run Code
docker run -it --gpus all -v /LOCAL_INPUT_FULL_PATH:/INPUTS -v /LOCAL_OUTPUT_FULL_PATH:/OUTPUTS --shm-size=250gb onealbao/hovernet_gca:1.1 bash -c "/app/script/run_pannuke_ndpi_40x_custom_batch_workers.sh 32 8 16"
docker run -it --gpus all -v /LOCAL_INPUT_FULL_PATH:/INPUTS -v /LOCAL_OUTPUT_FULL_PATH:/OUTPUTS onealbao/hovernet_gca:1.1 bash -c "/app/script/run_conic_ndpi_40x.sh"
Description
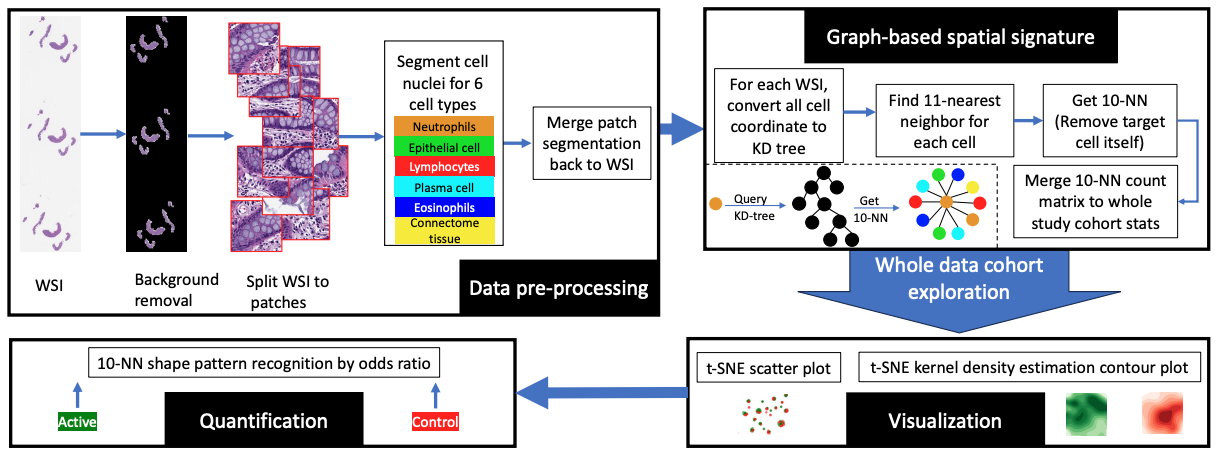
Crohn’s disease (CD) is a chronic and relapsing inflammatory condition that affects segments of the gastrointestinal tract. CD activity is determined by histological findings, particularly the density of neutrophils observed on Hematoxylin and Eosin stains (H&E) imaging. However, understanding the broader morphometry and local cell arrangement beyond cell counting and tissue morphology remains challenging. To address this, we characterize six distinct cell types from H&E images and develop a novel approach for the local spatial signature of each cell.
The contribution of this study is three-fold: (1)We propose a graph-based signature to represent a local arrangement signature for each cell. (2) We develop a comprehensive visualization workflow to identify the relationship of signature patterns among active CD and normal CD, and healthy control cohorts. (3) We present investigations from two research institutes with heterogeneous data acquisition, focusing on the rectum.
Reference
Cell Spatial Analysis in Crohn’s Disease: Unveiling Local Cell Arrangement Pattern with Graph-based Signatures
Shunxing Bao, Sichen Zhu, Vasantha L Kolachala, Lucas W Remedios, Yeonjoo Hwang, Yutong Sun, Ruining Deng, Can Cui, Yike Li, Jia Li, Joseph T Roland, Qi Liu, Ken S Lau, Subra Kugathasan, Peng Qiu, Keith T Wilson, Lori A Coburn, Bennett A Landman, Yuankai Huo
SPIE Medical Imaging 2024 (accepted)