This is the official implementation of Segment anything model (SAM) for digital pathology: Assess zero-shot segmentation on whole slide imaging
Source code repository
https://github.com/hrlblab/SAM_pathology
Get our docker image
Todo
Run code
Todo
Description
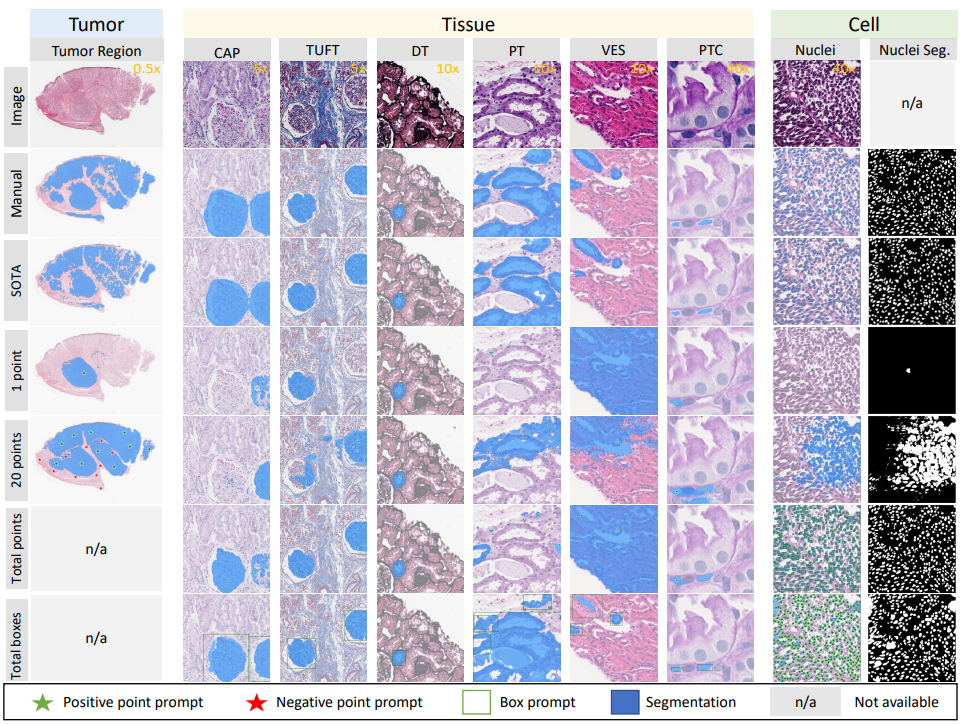
The segment anything model (SAM) was released as a foundation model for image segmentation. The promptable segmentation model was trained by over 1 billion masks on 11M licensed and privacy-respecting images. The model supports zero-shot image segmentation with various segmentation prompts (e.g., points, boxes, masks). It makes the SAM attractive for medical image analysis, especially for digital pathology where the training data are rare. In this study, we evaluate the zero-shot segmentation performance of SAM model on representative segmentation tasks on whole slide imaging (WSI), including (1) tumor segmentation, (2) non-tumor tissue segmentation, (3) cell nuclei segmentation.
The results suggest that the zero-shot SAM model achieves remarkable segmentation performance for large connected objects. However, it does not consistently achieve satisfying performance for dense instance object segmentation, even with 20 prompts (clicks/boxes) on each image. We also summarized the identified limitations for digital pathology: (1) image resolution, (2) multiple scales, (3) prompt selection, and (4) model fine-tuning.
Referece
Segment Anything Model (SAM) for Digital Pathology: Assess Zero-shot Segmentation on Whole Slide Imaging
Ruining Deng, Can Cui, Quan Liu, Tianyuan Yao, Lucas W. Remedios, Shunxing Bao, Bennett A. Landman, Lee E. Wheless, Lori A. Coburn, Keith T. Wilson, Yaohong Wang, Shilin Zhao, Agnes B. Fogo, Haichun Yang, Yucheng Tang, Yuankai Huo
MIDL2023 conference