This is the official implementation of MxIF Q-score: Biology-Informed Quality Assurance for Multiplexed Immunofluorescence Imaging.
Source code repository
https://github.com/MASILab/MXIF_Q_SCORE
Get our docker image
To do
Run MxIF Q-score
To do
Description
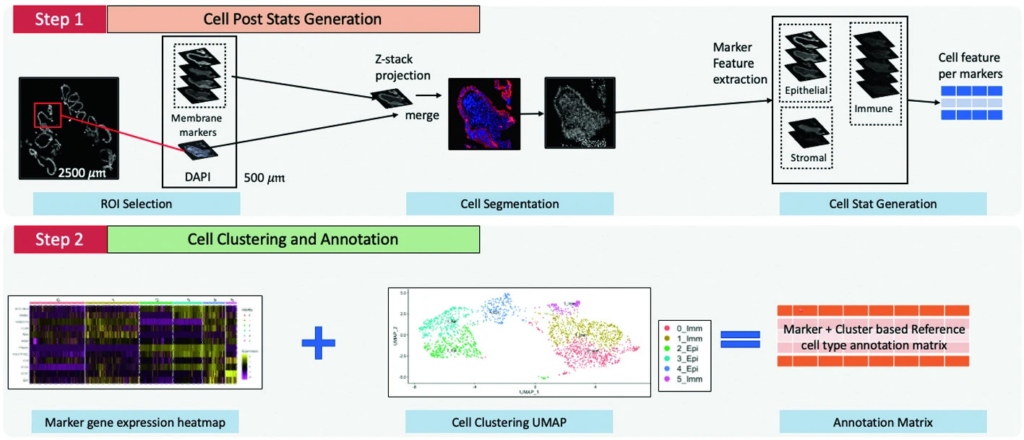
Medical image processing and analysis on whole slide imaging (WSI) are notoriously difficult due to its giga-pixel high-resolution nature. Multiplex immunofluorescence (MxIF), a spatial single-cell level iterative imaging technique that collects dozens of WSIs on the same histological tissue, makes the data analysis an order of magnitude more complicated. The rigor of downstream single-cell analyses (e.g., cell type annotation) depends on the quality of the image processing (e.g., multi-WSI alignment and cell segmentation). Unfortunately, the high-resolutional and high-dimensional nature of MxIF data prevent the researchers from performing comprehensive data curations manually, thus leads to misleading biological findings.
In this paper, we propose a scoring framework, MxIF Q-score that integrates an automatic image segmentation pipeline and a single-cell clustering method to conduct MxIF image processing quality guidance. The contribution of this paper is three-fold: (1) An biological knowledge-informed metrics, MxIF Q-score, is proposed for an objective and automatic QC on high-resolution and high-dimensionality MxIF data. (2) Comprehensive experiments on different MxIF Q-score thresholds have been conducted that demonstrate the usefulness analysis of the MxIF Q-score framework. (3) To our knowledge, MxIF Q-score is the first quantitative and objective metrics to meet the emergent needs of curating MxIF data.
Reference
MxIF Q-score: Biology-informed Quality Assurance for Multiplexed Immunofluorescence Imaging
Shunxing Bao, Jia Li, Can Cui, Yucheng Tang, Ruining Deng, Lucas W. Remedios, Ho Hin Lee, Sophie Chiron, Nathan Heath Patterson, Ken S. Lau, Lori A. Coburn, Keith T. Wilson, Joseph T. Roland, Bennett A. Landman, Qi Liu, Yuankai Huo
Medical Optical Imaging and Virtual Microscopy Image Analysis (MOVI) @ MICCAI 2022